Transcriptomic Sequencing
Whole transcriptome sequencing, also called total RNA-Seq, is the ideal approach to gain a complete picture of the entire transcriptomes of the organism of interest. It detects coding plus multiple forms of noncoding RNA. Total RNA-Seq can accurately measure gene and transcript abundance, and identify known and novel features of the transcriptome. By looking at the whole transcriptome, researchers are able to determine global expression levels of each transcript, identify exons, introns and map their boundaries. In addition, RNA-Seq can be used for the identification of splicing variants. Since ribosomal RNA (rRNA) accounts for as much as 98% of the transcriptome, the rRNA is often depleted in order to optimize the number of reads covering the actual RNA of interest.
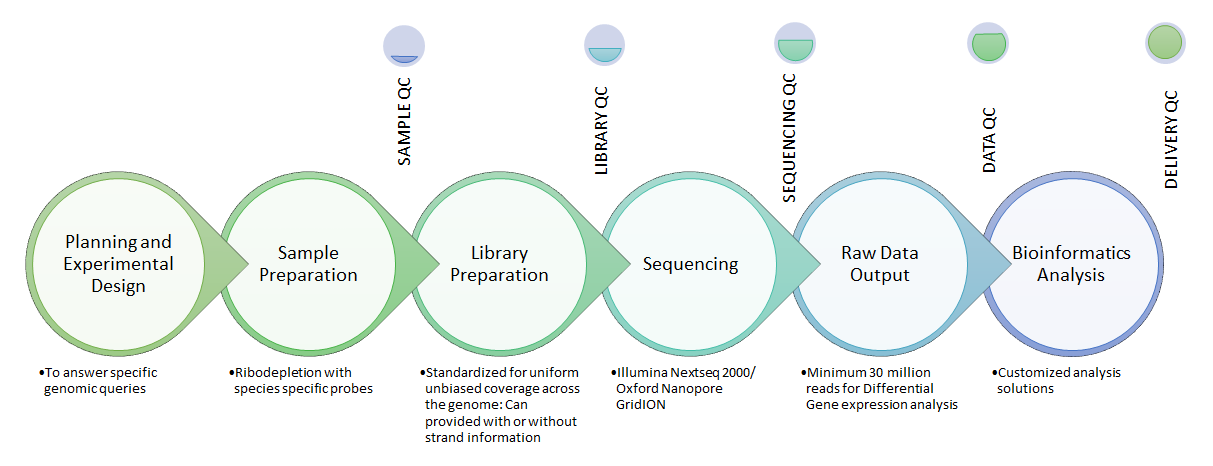
We at National Genomics Core-CDFD have meticulous quality control in each step of the sequencing project. Experts with PhD and International experience are using top-notch technologies to carefully plan and generate high quality, meaningful data.
Transcriptome sequencing is used to reveal the presence, quantity and structure of RNA in a biological sample under specific conditions. Compared to hybridization-based RNA quantification methods such as microarray analysis, sequencing-based transcriptome detection can quantify gene expression with low background, high accuracy and high levels of reproducibility within a large dynamic range. In addition, transcriptome sequencing does not require an existing genome sequence and can detect mutations, splice variants and fusion genes that cannot be detected by microarrays.
Messenger RNA-Seq or mRNA-Seq is a targeted RNA-Seq protocol that enriches for all polyadenylated (poly-A) transcripts of the transcriptome. mRNA-Seq is a method used in studying transcription in disease states as well as expression in variety of research based applications. Only around 1-2% of the entire transcriptome is comprised of poly-A tailed RNA, the coding part of the genome. By targeting mRNA, sequencing depth is improved as resources are dedicated to the sequencing of coding genes. This makes identifying rare variants and low expressed mRNA transcripts easier.
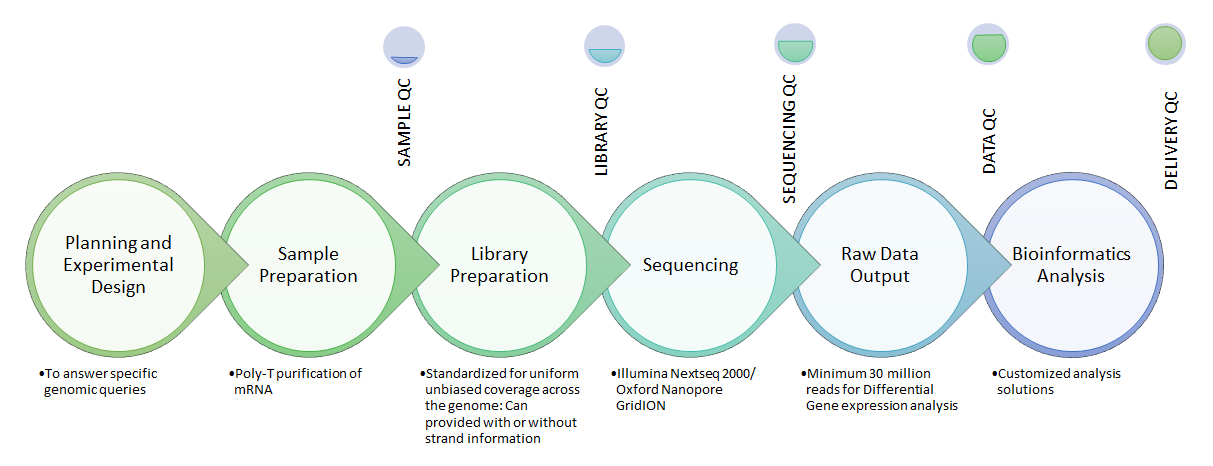
We at National Genomics Core-CDFD have meticulous quality control in each step of the sequencing project. Experts with PhD and International experience are using top-notch technologies to carefully plan and generate high quality, meaningful data.
Transcriptome sequencing is used to reveal the presence, quantity and structure of RNA in a biological sample under specific conditions. Compared to hybridization-based RNA quantification methods such as microarray analysis, sequencing-based transcriptome detection can quantify gene expression with low background, high accuracy and high levels of reproducibility within a large dynamic range. In addition, transcriptome sequencing does not require an existing genome sequence and can detect mutations, splice variants and fusion genes that cannot be detected by microarrays.
Small RNAs are a type of non-coding RNA (ncRNA) molecule that are less than 200nt in length. They are often involved in gene silencing and post- transcriptional regulation of gene expression. Small RNA sequencing is used to discover novel small RNAs, examine the differential expression of all small RNAs and to characterize variations with single-base resolution.
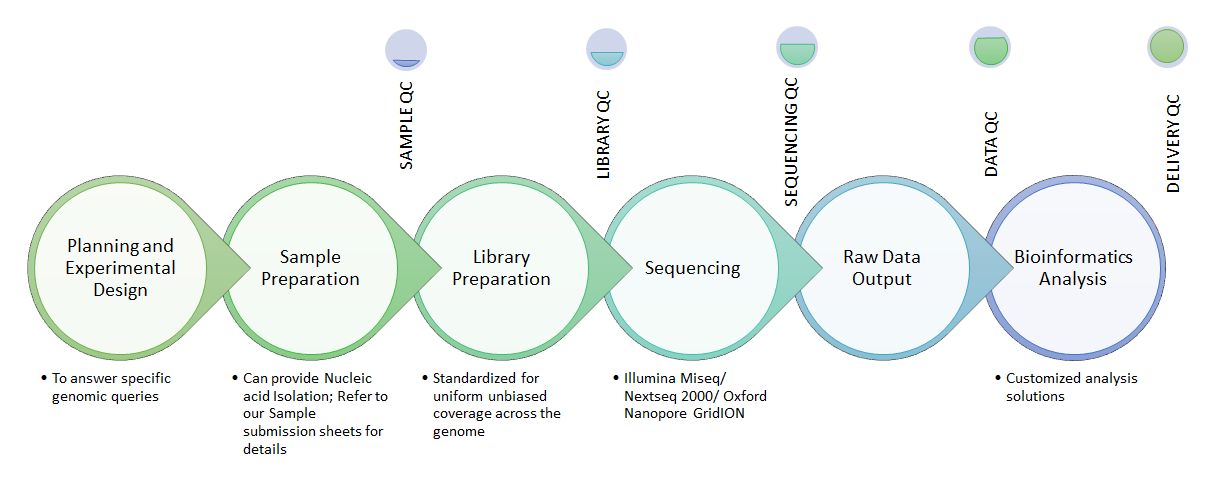
We at National Genomics Core-CDFD have meticulous quality control in each step of the sequencing project. Experts with PhD and International experience are using top-notch technologies to carefully plan and generate high quality, meaningful data.
In 2015, Oxford Nanopore announced the ability to sequence RNA strands on the MiniION™ and PromethION™ without needing to convert to double stranded DNA. Direct sequencing of RNA eliminates biases associated with transcription and provides a more accurate measure of transcript profiles. Modified RNA bases can be directly measured, whereas with cDNA conversion they are lost
We at National Genomics Core-CDFD have meticulous quality control in each step of the sequencing project. Experts with PhD and International experience are using top-notch technologies to carefully plan and generate high quality, meaningful data.